Regulating Gene Expression (College Board AP® Biology): Study Guide
Introduction to regulating gene expression
The genes expressed by a cell determine a cell's structure and function; this means that it is essential for cells to regulate their gene expression
There are several ways in which gene expression can be regulated:
Regulatory proteins: proteins that bind to specific base sequences in DNA to switch gene expression on or off, e.g. transcription factors
Regulatory sequences: sequences of DNA to which regulatory proteins bind, e.g. promoters
Constitutive vs inducible genes:
Constitutive genes are always expressed (e.g. housekeeping genes needed for basic cell function).
Inducible genes are expressed only when required (e.g. enzymes in the lac operon).
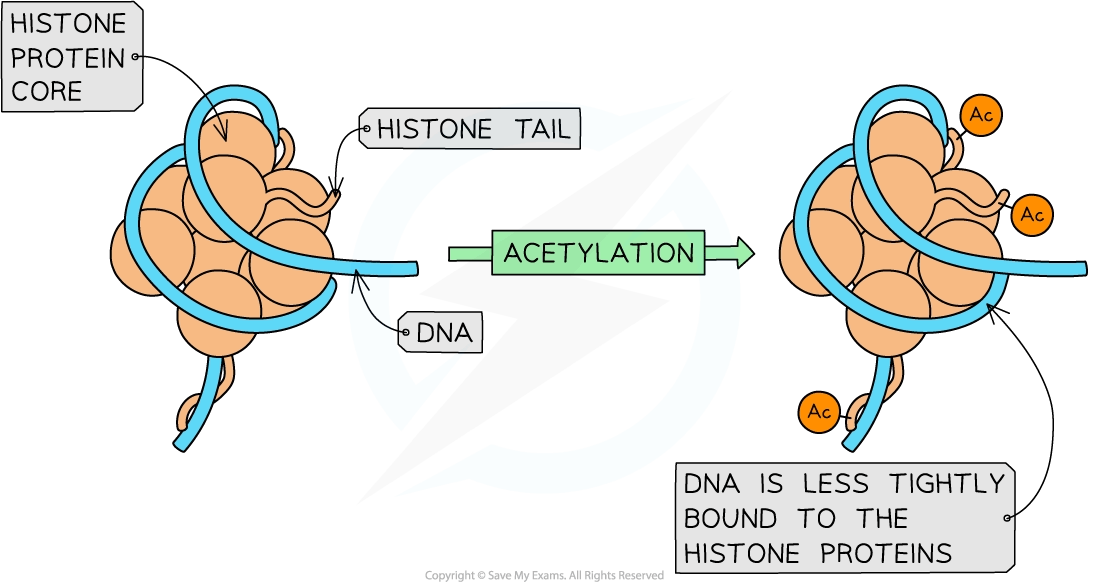
Epigenetic changes: reversible modifications to DNA or histones, e.g.:
methylation of DNA: methyl groups can be added to DNA, reducing transcription rates
acetylation of histones: acetyl groups are added to histones, increasing transcription rates
Small interfering RNA (siRNA): single-stranded molecules of RNA that bind to complementary regions of mRNA, preventing them from being translated
Regulation and phenotype: The combination and level of gene expression determine cell and organism phenotype
Cell differentiation: occurs when genes for tissue-specific proteins are expressed (e.g. muscle vs nerve cells)
Sequential gene expression: during development, transcription factors activate cascades of genes in a set order
Phenotype outcomes: both the function of gene products and the amount produced influence traits
Coordinate regulation
Regulatory sequences are stretches of DNA that control gene expression
Their location relative to the gene(s) they control determines how they function:
Near the gene (cis-regulatory elements): directly influences transcription initiation
Upstream, downstream, or within introns: can act as enhancers or silencers
Grouped together: allows coordinated regulation of multiple genes
Coordinate regulation in prokaryotes – operons
An operon is a cluster of genes under the control of a single promoter and operator
The regulatory sequence (operator) is directly upstream of the genes
Ensures all the genes in the operon are transcribed together as one mRNA, so proteins needed for the same function are made at the same time
Prokaryotes regulate operons in inducible or repressible systems
Inducible operons: These are normally off and will be switched on by an inducer (e.g. lac operon activated by lactose)
Repressible operons: These are normally on and will be switched off when the end product is abundant (e.g. trp operon repressed by tryptophan)
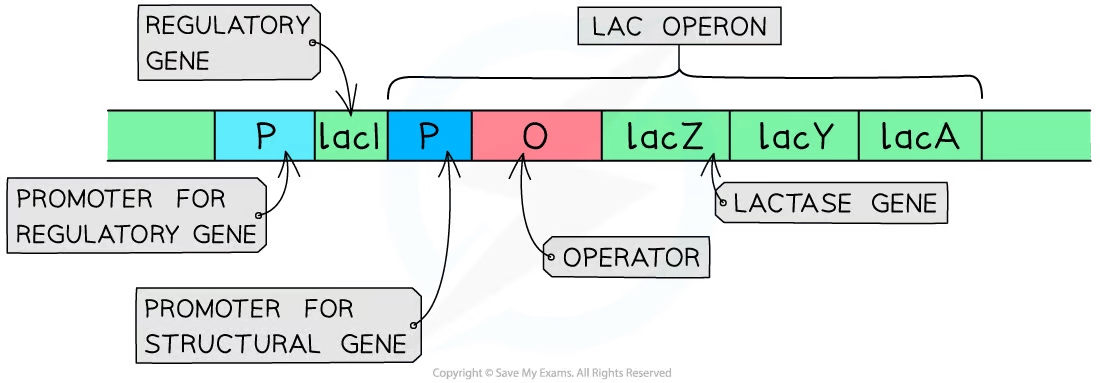
Inducible system example: the lac operon
The lac operon in bacteria controls the genes for lactose-digesting enzymes.
It is inducible—normally off, but switched on when lactose is present
Components of the lac operon include:
Structural genes (lacZ, etc.) code for enzymes that digest lactose
Regulatory genes code for regulatory proteins that switch the expression of structural genes on and off
Regulatory sequences include the promoter (RNA polymerase binding site) and operator (repressor binding site)
The lac operon is an example of an inducible system in prokaryotes
The effect of lactose
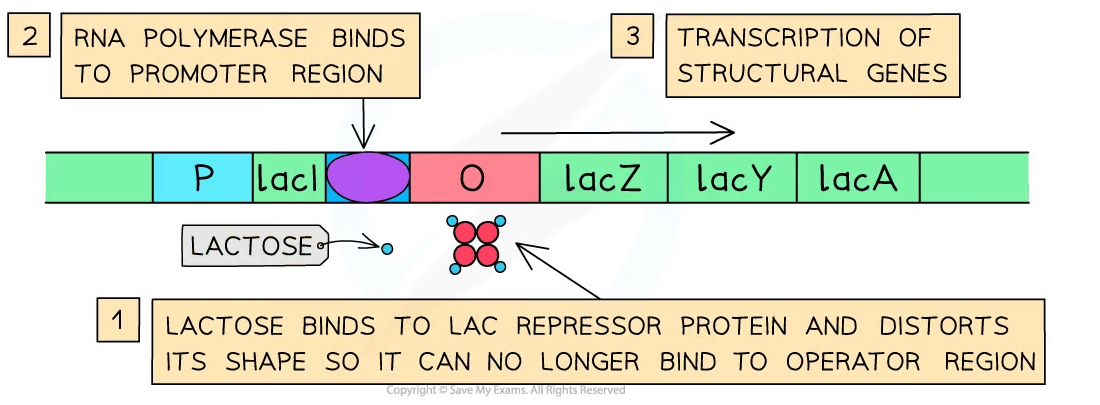
Without lactose:
The repressor binds to the operator, blocking RNA polymerase
Structural genes are not transcribed, and no enzymes are made
With lactose:
Lactose binds to the repressor, preventing it from binding to the operator.
RNA polymerase transcribes the structural genes, enzymes are produced, and lactose is broken down
Repressible system example: the trp operon
The trp operon controls enzymes for tryptophan synthesis
It is repressible, meaning it is normally on, but will be switched off when tryptophan (trp) is abundant
Without tryptophan:
The repressor cannot bind the operator
RNA polymerase transcribes the structural genes, and tryptophan is produced
With tryptophan:
Tryptophan binds the repressor, enabling it to block the operator
RNA polymerase is prevented from transcribing, so synthesis stops and energy is conserved
Coordinate regulation in eukaryotes – shared transcription factors
Genes are usually dispersed across different chromosomes rather than clustered
These genes can still be coordinately regulated if they share regulatory sequences that bind the same transcription factor
For example, heat-shock genes all contain a similar regulatory element, and a single transcription factor activates them together during stress
Regulatory sequences are stretches of DNA that control transcription
They can be found:
Upstream (before the gene)
Downstream (after the gene)
Within introns (non-coding regions inside the gene)
Some regulatory sequences act as enhancers: binding sites for transcription factors that increase transcription
Even if enhancers are far away, the DNA can loop around so that transcription factors bound at the enhancer physically touch the promoter region
This contact helps RNA polymerase bind to the promoter and start transcription
Comparison of coordinate regulation
Feature | Prokaryotes (operons) | Eukaryotes (transcription factors) |
|---|---|---|
Gene arrangement | Clustered in operons | Dispersed across genome |
Regulatory sequence | Promoter + operator next to the genes | Promoters, enhancers, silencers → may be far from genes |
Mechanism of coordination | Single promoter controls all genes | Shared transcription factor activates different genes |
Example | lac operon (inducible) | Heat-shock protein genes |
Examiner Tips and Tricks
You will not be expected to remember specific examples of inducible and repressible systems, but you need to understand how these systems function and be able to apply your knowledge to unfamiliar examples that may be presented in an exam.
Transcription factors
Transcription factors are proteins that bind to regulatory DNA sequences (such as promoters or enhancers) to control gene expression
Their action influences an organism’s phenotype by determining whether a gene is transcribed
Promoters and transcription factors
Promoters are DNA sequences located immediately upstream of a gene
They are the binding site for RNA polymerase and mark where transcription begins
Every gene needs a promoter for basic transcription initiation, usually close to the transcription start site
Transcription factors are proteins that recognise and bind to promoter (or nearby) sequences
They control how easily RNA polymerase can bind and therefore, regulate whether transcription starts
Activators help recruit or stabilise RNA polymerase binding, increasing transcription
Repressors/negative regulators block RNA polymerase from binding or moving along the DNA, reducing or preventing transcription
In eukaryotes, multiple transcription factors often work together in complexes to finely tune expression; in prokaryotes, fewer factors are involved, but the principle is the same

Enhancers and transcription factors
Enhancers are regulatory DNA sequences that increase the rate of transcription
Unlike promoters, they can be located far upstream, downstream, or within introns of the gene they regulate
Transcription factors (activators) bind to enhancer sequences
The DNA can loop around so that these activators come into physical contact with the promoter region
This interaction helps recruit and stabilise RNA polymerase at the promoter, making transcription more likely to start
Enhancers, therefore, act as boosters, fine-tuning gene expression depending on the presence of specific transcription factors

Unlock more, it's free!
Did this page help you?